Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from June, 2012
Posted by
Varun C N
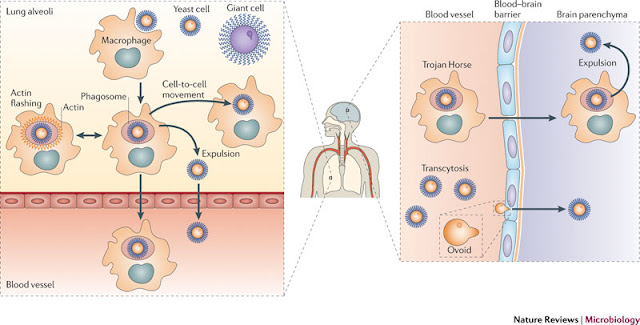
Clash of the titan- Survival strategy for Cryptococcus
- Get link
- Other Apps

Posted by
Varun C N
Pore-fection aims perfection
- Get link
- Other Apps
Posted by
Varun C N
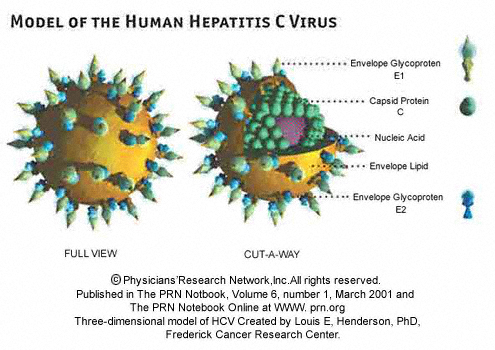
The mi-RNA game of HCV
- Get link
- Other Apps
Posted by
Varun C N
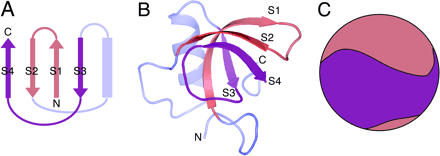
Hydrophobins- A snapshot
- Get link
- Other Apps