Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from April, 2013

Posted by
Varun C N
Controlling malaria- Perspectives
- Get link
- Other Apps

Posted by
Varun C N
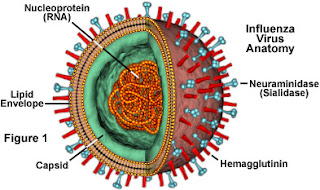
H7N9- Follw up post
- Get link
- Other Apps
Posted by
Varun C N
H7N9 in News
- Get link
- Other Apps

Posted by
Varun C N
Resistance Detection- Faster the better
- Get link
- Other Apps