Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from January, 2014
Posted by
Varun C N
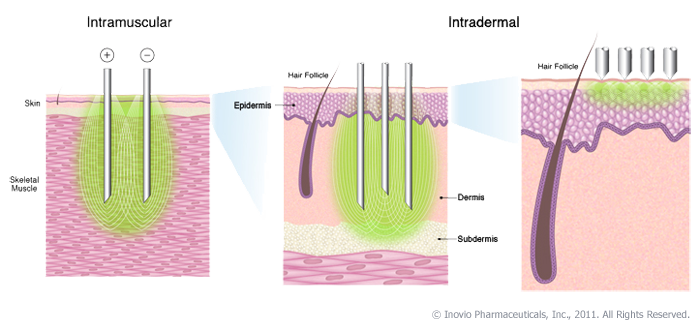
NextGen vaccination: Needle free devices, Interleukin adjuvants
- Get link
- Other Apps

Posted by
Varun C N
Clostridium difficile on a note
- Get link
- Other Apps
Posted by
Varun C N
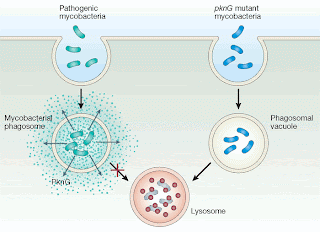
PDIM: Yet another virulence factor of TB
- Get link
- Other Apps

Posted by
Varun C N
Bacterial Persistence- On a Note
- Get link
- Other Apps