Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from January, 2016
Posted by
Varun C N
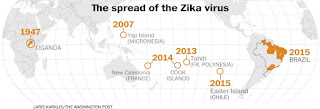
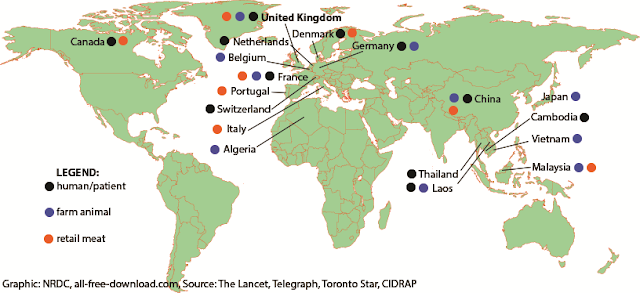
MCR-1 gene: Update
- Get link
- Other Apps

Posted by
Varun C N
Who wins by count: Man Vs Microbiome
- Get link
- Other Apps

Posted by
Varun C N
Lab series# 9- Pulsed field gel electrophoresis
- Get link
- Other Apps

Posted by
Varun C N
Blogger's Desk #9: For Science Blogger's
- Get link
- Other Apps