Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from February, 2016

Posted by
Varun C N
Guest Post: What it is to be a part of industry?
- Get link
- Other Apps

Posted by
Varun C N
BtB#6: H pylori Urease
- Get link
- Other Apps
Posted by
Varun C N
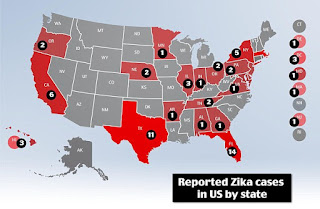
Zika Virus spread 2016- Update
- Get link
- Other Apps
Posted by
Varun C N
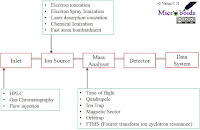
Lab Series# 10: MALDI TOF in Microbial diagnostics
- Get link
- Other Apps