Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from February, 2014
Posted by
Varun C N
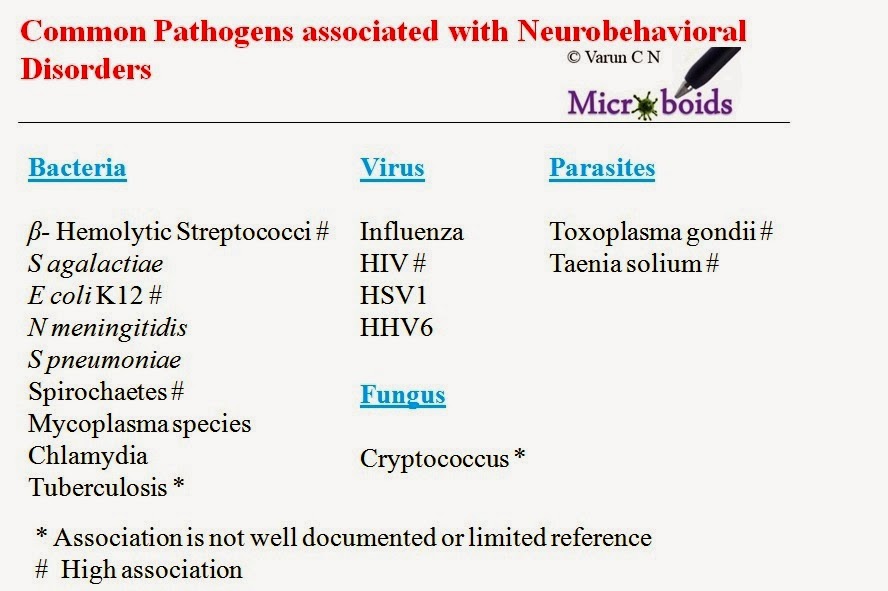
Schizophrenia- Ever thought of Microbial involvement?
- Get link
- X
- Other Apps

Posted by
Varun C N
Protein A, G, A/G, L and Protein M
- Get link
- X
- Other Apps

Posted by
Varun C N
Clustered Regularly Interspaced Short Palindromic Repeats- CRISPR
- Get link
- X
- Other Apps