Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from May, 2016

Posted by
Varun C N
The link between Brain and Microbiome
- Get link
- X
- Other Apps
Posted by
Varun C N
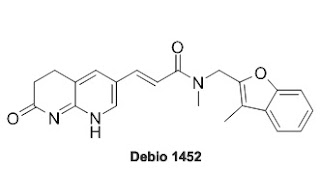
An Antibiotic that doesn't effect Gut Microbiome.
- Get link
- X
- Other Apps

Posted by
Varun C N
3000 Microbes: Different looking life tree
- Get link
- X
- Other Apps