Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from June, 2016

Posted by
Varun C N
BtB#7: Immune cells of Central Nervous System
- Get link
- X
- Other Apps

Posted by
Varun C N
NGS for Microbial Diagnostics
- Get link
- X
- Other Apps

Posted by
Varun C N
VDPV in Sewer sample- Implications
- Get link
- X
- Other Apps
Posted by
Varun C N
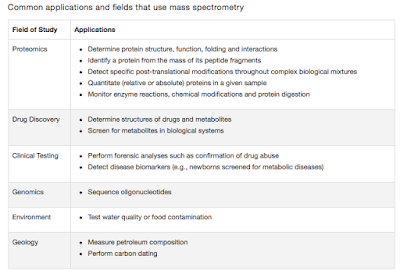
Lab series# 13: Mass Spectrometry Instrumentation Principles
- Get link
- X
- Other Apps
Posted by
Varun C N
Lab Series# 12: Cryo-Preservation of cells
- Get link
- X
- Other Apps

Posted by
Varun C N
Totally Drug Resistant Tuberculosis is actually XDR Plus
- Get link
- X
- Other Apps

Posted by
Varun C N
First case of MCR-1 in US
- Get link
- X
- Other Apps
