Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from January, 2017
Posted by
Varun C N
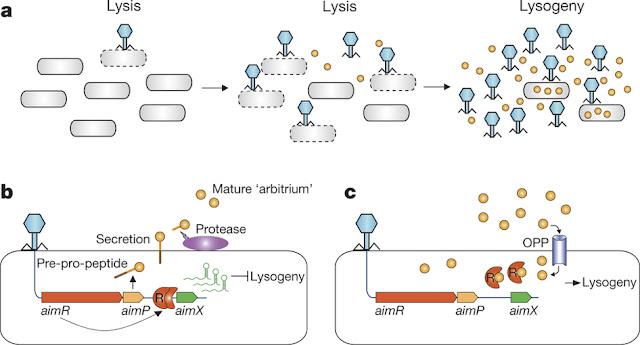
Speaking Phage language
- Get link
- X
- Other Apps
Posted by
Varun C N
Report of a Klebsiella pneumoniae strain resistant to 26 antibiotics
- Get link
- X
- Other Apps

Posted by
Varun C N
Blocking the CRISPR
- Get link
- X
- Other Apps

Posted by
Varun C N
Predatory bacteria in news
- Get link
- X
- Other Apps