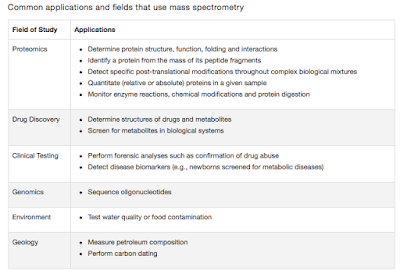
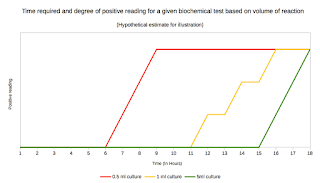
Clustered Regularly Interspaced Short Palindromic Repeats- CRISPR
Greetings
I was going through the statistics of this blog, in context of what is read and discussed the most. I presume antibiotics, HIV and Influenza are the most read and searched topics. It reflects very much on how much the current science craves for a good antibiotic. From my reading (If i understood it correct), we are never going to have the perfect antibiotic, but maybe we will have better. For now, i will talk about a yet another basic of molecular bacteriology, the CRISPR system.
A long time ago, I had written a very basic post about CRISPR system. This probably is the right time to revisit the topic. CRSIPR stands for Clustered Regularly Interspaced Short Palindromic Repeats. CRSIPR genetic system is always in association with a set of functional regulators called as cas (CRSIPR associated) genes. Together they are referred as CRSIPR/Cas system. Classically, they are a intracellular microbial defense system, (a type of microbial nuclease) involved in defense against invading phages. The system can be considered as analogous to the RNAi mechanism. A target sequence remembers the sequence of RNA to be attacked, which binds and cleaves the target nucleic acid. Based on the structure, activity and mechanism of functioning, it is classified into 3 types and several sub types.
 |
| Fig 1: Summary of CRISPR Cas System. |
 |
| Fig 2: Basic Structure of CRISPR system |
The basic structure of CRISPR Cas genome consists of a leader sequence, Cas enzymes, repeating sequence, interspersed with spacer sequences. The basic sequence of events are acquisition, expression and interference. The acquisition of phage DNA happens during the initial phage infection. It is not entirely known as to how this functions. The bottom line is randomly sequence are captured and integrated to the CRISPR system as a spacer sequence. Of note, we can have a pretty good idea of the history of phage infection of the bacteria by knowing the sequence of these spacers. This sequence forms the memory of immunity. The spacer is usually about 30 bp in length. Every time a new spacer is added a repeat unit is also attached creating space for the next integration. The setup of spacers which is due to processed is called as Proto-spacer-adjacent motifs (PAM's). Protospacers are defined as short sequences (~20 bp) of known foreign DNA separated by a short palindromic repeat and kept like a record against future encounters.
The second step is Expression. The repeat-spacer array is processed to yield small CRISPR RNAs (crRNAs) that contain a full or partial spacer sequence. This crRNA acts as the guide (referred as guide RNA or gRNA in artificial system). This in itself is a multi- step process with formation of intermediates including trans-activating crRNA (tracrRNA). For a detailed step by step process, see here.
 |
| Fig 3: Basic Steps in CRISPR Cas functioning |
A little bit of clarification here. Some literature suggests 2 steps- A the highly conserved "Information processing subsystem", which refers to Acquisition (Adaptation), and the "executive" subsystem, which includes the expression and interference stages. This is to demarcate the enzymes involved. The first step uses enzymes that are almost universal in the bacterial population (Cas1 and Cas2), whereas the executive system depends on the organism under question.
The final step is interference. Once a complete mature crRNA which fits by base pairing to the viral genome that maybe present, is formed, it triggers a series of events (depending on the type of subsystem). The final outcome being degradation of DNA or RNA.
I want to make an important point here. To have immunity against more phages demands more spacer sequences. Though theoretically you are allowed to have as many sequences as you wish to have, practically, the size itself becomes a limiting factor. Research suggests that, this problem is possibly circumvented by deleting very old memories which is not actively encountered. And that lets the bacteria be immune- erased against that phage. The second important thing to be noted is that the "Nucleic Acid guided attack" is highly sequence specific. This means, a very small change in the phage genome can lead to significant non recognition, defeating the whole purpose. This is how exactly a phage evolves to evade the immunity put up by the bacteria.
The CRISPR specificity can be used as an advantage point in gene editing. The same editing system can be turned against the bacteria itself, by some neat tricks. It is well known that intentional targetting and disruption of bacterial genome can be very deleterious. A paper by Gomaa etal published in mBio, used this understanding to design a CRISPR system (type I-E CRISPR-Cas system) that could attack E coli selectively in a proof of concept model experiment. The authors suggest that this is an excellent antibiotic design. If a feasible mode of delivery can be invented, this may serve as an important selective anti bacterial strategy.
Signing of with a take home message. The Zinc finger nuclease and CRSIPR technology (both inspired from natural systems) has come up as an excellent nucleic acid editing tool. But, there is a still lot to be studied about CRISPR, as many of the participant enzymes, and the pathways of this system is not well understood. There is always scope for more.
I want to make an important point here. To have immunity against more phages demands more spacer sequences. Though theoretically you are allowed to have as many sequences as you wish to have, practically, the size itself becomes a limiting factor. Research suggests that, this problem is possibly circumvented by deleting very old memories which is not actively encountered. And that lets the bacteria be immune- erased against that phage. The second important thing to be noted is that the "Nucleic Acid guided attack" is highly sequence specific. This means, a very small change in the phage genome can lead to significant non recognition, defeating the whole purpose. This is how exactly a phage evolves to evade the immunity put up by the bacteria.
The CRISPR specificity can be used as an advantage point in gene editing. The same editing system can be turned against the bacteria itself, by some neat tricks. It is well known that intentional targetting and disruption of bacterial genome can be very deleterious. A paper by Gomaa etal published in mBio, used this understanding to design a CRISPR system (type I-E CRISPR-Cas system) that could attack E coli selectively in a proof of concept model experiment. The authors suggest that this is an excellent antibiotic design. If a feasible mode of delivery can be invented, this may serve as an important selective anti bacterial strategy.
Signing of with a take home message. The Zinc finger nuclease and CRSIPR technology (both inspired from natural systems) has come up as an excellent nucleic acid editing tool. But, there is a still lot to be studied about CRISPR, as many of the participant enzymes, and the pathways of this system is not well understood. There is always scope for more.
Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, & Koonin EV (2011). Evolution and classification of the CRISPR-Cas systems. Nature reviews. Microbiology, 9 (6), 467-77 PMID: 21552286
Bikard D, & Marraffini LA (2013). Control of gene expression by CRISPR-Cas systems. F1000prime reports, 5 PMID: 24273648.
Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, & Beisel CL (2014). Programmable Removal of Bacterial Strains by Use of Genome-Targeting CRISPR-Cas Systems. mBio, 5 (1) PMID: 24473129
Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, & Beisel CL (2014). Programmable Removal of Bacterial Strains by Use of Genome-Targeting CRISPR-Cas Systems. mBio, 5 (1) PMID: 24473129


Comments
Post a Comment