Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from April, 2015

Posted by
Varun C N
Blogger's Desk #6: Spark of Ethical Debate on editing the embryonic genome
- Get link
- X
- Other Apps
Posted by
Varun C N
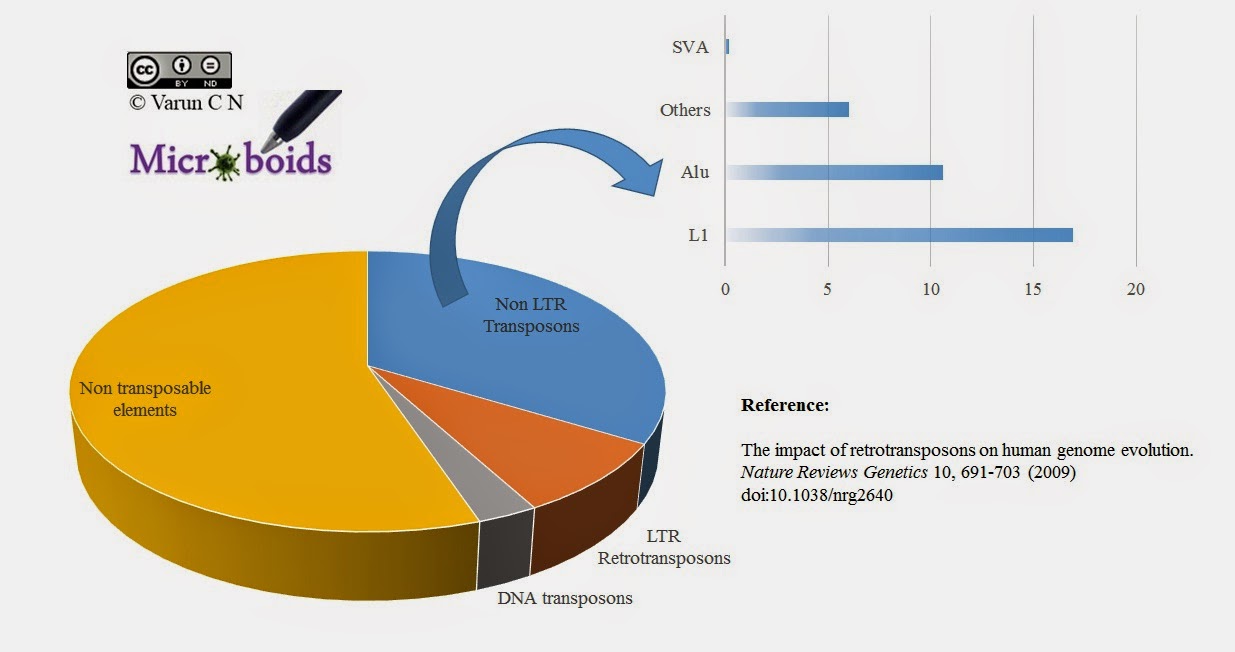
Retroviral protection
- Get link
- X
- Other Apps

Posted by
Varun C N
Lab Series #3: Flow Cytometry
- Get link
- X
- Other Apps
Posted by
Varun C N
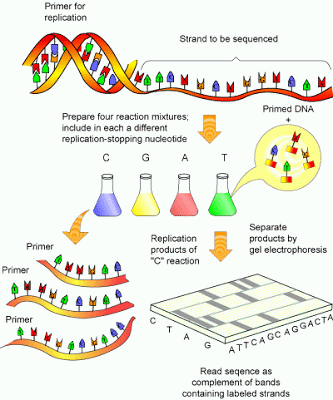
Lab Series #2: DNA sequencing
- Get link
- X
- Other Apps

Posted by
Varun C N
Lab Series #1: Luminescence
- Get link
- X
- Other Apps