Lab Series #2: DNA sequencing
Greetings,
One of the most important techniques that is relied upon in the world of life science is to study the sequence of gene of interest. The technique to determine the sequence of bases in DNA is called DNA sequencing which is the central part of current genetic technologies. The classical methods available for sequencing DNA include
- Sanger’s sequencing method (dideoxynucleotide method)
- Direct PCR Pyrosequencing
- Maxam and Gilbert sequencing
Sanger’s method
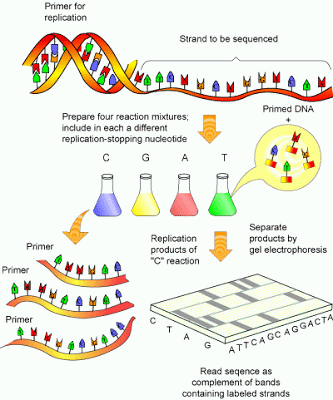 |
| Fig 1: Sanger sequencing. Source |
The DNA is denatured by heat or more traditionally inserted and cloned into a vector M13 (which is naturally single stranded). The DNA is extracted and the reaction mixture is then divided into four aliquots. The tube A contains all the 4 nucleotides and 2’, 3’-dideoxyadenosine triphosphates (ddATP). Similarly tube T contains all nucleotides and ddATT and so on. The dideoxynucleotide doesn’t possess a 3’ end and so will terminate the synthesis, since polymerase an add nucleotides only to the 3’-end.
The incorporation of ddNTP will be a random event, the reaction producing molecules of various lengths culminating in the same ddNTP. The reaction products are then run using an electrophoresis method (commonly used is polyacrylamide gel method). The position of the various bands of each ddNTP will be the indication of sequence (See Fig 5). Under ideal conditions sequences up to about 300 bases in length can be read from as single gel run.
Direct PCR Pyrosequencing
 |
| Fig 2: Pyrosequencing Source |
This is a sequencing method were a PCR template is hybridized to an oligonucleotide and incubated with a DNA polymerase, ATP sulphurylase, luciferase and apyrase. During the reaction the first of the 4 dNTP are added and if incorporated release pyrophosphate (PPi). The ATP sulphurylase converts the PPi to ATP. This ATP can now convert the luciferin to oxyluciferin to generate light. The overall reaction is
dNTP + ATP sulphurylase +luciferin ---------------------------> light
This is followed by another round of addition of dNTP. The resulting program can b used for the analysis of sequence. The method described represents a very fast sequencing result with good potential to be automated. It also provides highly precise and accurate analysis. Also it avoids the problems of gel electrophoresis.
Maxam and Gilbert sequencing:
The DNA is radiolabelled with 32P at the 5’ ends of each strand and the strands denatured separated and purified to give a population of labeled strands for the sequencing reactions. The next step is a chemical modification of the bases in the DNA strand. The modified bases are then removed from their sugar groups and the strands cleaved at these positions using the chemical piperidine. This will create a set of fragments known as nested fragments. It is then analysed for products as in Sanger’s method.
Automated fluorescent DNA sequencing:
The method is similar to that of the Sanger’s method. Here a specific fluorescent dye terminator (instead of ddNTP) is used in a single reaction cuvette. This allows a single gel column to be run. The sequence can be read using a laser (Light amplification by stimulated emission of radiation) and detecting the fluorescence which instead tells about the sequence. The method can be connected to various to various other bioinformatics software’s that allows high data collection.
Needless to say, the first generation technology "Sanger sequencing" method is still considered as a gold standard in resolving issues such as looking into SNP (single nucleotide polymorphism). With sequencing methods automated and access to better technology, the human genome project which was once a mammoth project is now a more feasible technology. But then the method in common laboratory remains cost prohibitive. So science seeks newer methods that can bring down the cost to less than 1000$ per human genome.
Many new techniques that rely on the basic DNA replication mechanisms are now introduced by the companies. Below I will discuss a couple since it has become too common now.
SMRT sequencer:
SMRT (pronounced as smart), stands for Single Molecule Real Time sequencers. This technology is a product of Pacific Biosciences corporation. The technology uses the same biological process that a natural system uses.
 |
| Fig 3: Phi29 polymerase. Source |
The first requirement is a DNA polymerase. The polymerase used here is φ29 polymerase. This polymerase is derived from a bacteriophage Φ29. This phage is a native attacker of B. subtilis. This polymerase has exceptional strand displacement and an inherent 3´-5' proofreading exonuclease activity. Its exceptional qualities have been useful in genetic studies. The enzyme is obtained in industrial quantities by cloning the gene to an E coli.
The second component of this technology are the special fluorescent nucleotides. Nucleotides are the basic structural units of DNA or RNA. In this case the 4 nucleotides- A, T, C, G are labelled with different fluorescent colors. The speciality is that they are γ-labelled dNTPs. Labeling at the terminal nucleotide is done on purpose. In a normal replication process, DNA polymerase cleave the α-β-phosphoryl bond upon incorporating a nucleotide into DNA, releasing the pyrophosphate leaving group and attached fluorescent label. This means when cleaved the γ- labelled fluorescent molecule is free to move out without being effected.
Of note, the fluorophore is attached to the nucleotide by using linkers. This attachment is cleavable, with chemicals so that the dye can be detached from the DNA after it has been detected. This serves to remove the noise in detection and thus enhance the assay. One important thing of note is that extension of the triphosphate moiety to four and five phosphates can increase incorporation efficiency (Reference). However, to the best of my understanding this idea has not been used in the technology.
 |
| Fig 4: ZMW cells. Source |
Third component of this system is the reaction chamber embedded in a chip. The chip is a glass cover slip with aprox 100 nm-thick layer of aluminum deposited on top of it. In this plate is an array of cylindrical wells each 70 nm–100 nm in diameter. The aluminum is chemically treated so that polymerase molecules will stick to the glass at the bottom of each well rather than the sides of the wells. Each well is designed to hold one polymerase molecule in it. The cover glass at the bottom is designed for feasibility of imaging.
The above mentioned special reaction cell is referred otherwise as ZMW (Zero mode waveguides). These cells are product of nano technology. The reaction chamber holds not more than a few atto- or zeptoliters. This is in the range of 10-18 to 10-21.They call it the microfluidics. But I think I better call it zeptofluidics. This permits use of extreme low sample volumes.
A problem often encountered in the whole genome sequencing is the limitedness of sequencing very large sequences. It is just not practical to sequence the whole set of chromosomes or even a single chromosome in one read due to technical constraints. The fastest method around the problem is a "shotgun" approach. This method breaks up the whole genome to fragments and then sequences each bits. Then the sequences are realigned using a computer program using unique matches.
So, this is how the technology works. The first step is to prepare the sample. The genomic DNA is obtained, and broken into fragments. Each fragmented DNA is then incorporated into a reaction chamber. The reaction starts with DNA polymerase which unwinds the DNA and incorporates the correct nucleotide. The γ-phosphate is released along with the fluorescent dye. Since the reaction can hold only one molecule of dNTP, in one of the four colors a binary value of 1 or 0 is generated. This implies the graph plot will show square peaks rather than the usual triangular peaks we are used to.
By simultaneously running a very large set of reaction in a complete chip, the full genome is sequenced at a very high speed. The method also avoids the requirement of running a gel and the sequence is obtained in real time. Since the signal generated is cleaved and released after each binding step, this reduces the otherwise background noise. Remember, reaction can hold only one molecule of dNTP.
As a proof of concept, the company has sequenced E coli O104:H4 strain with an accuracy of 99.9% and some sources claiming it to be 99.9999% accuracy. This level of accuracy is unheard of any other 1st or 2nd generation sequencers.
Dr. Schadt comments "The ability to sequence the outbreak strain with reads averaging 2,900 base pairs and our longest reads at over 7,800 bases, combined with our circular consensus sequencing to achieve high single molecule accuracy with a mode accuracy distribution of 99.9%, enabled us to complete a PacBio-only assembly without having to construct specialized fosmid libraries, perform PCR off the ends of contigs, or other such techniques that are required to get to similar assemblies with second generation DNA sequencing technologies." And it took them less than 8 hrs on average to complete sequencing.
Ion Proton Sequencer
Next lets discuss about another sequencer: Ion Proton Sequencer. The technology comes to the laboratory bench from Life technologies. The technology is vaguely referred as Semi conductor sequencing. The system was unpacked to the world of science in January 2012 and testified of $1,000, full genome sequence in a single day. Oh and by the way, Jonathan Rothberg is considered as the innovator of this technology.
 |
Photo 1: Ion Proton™ Sequencer
|
The technology works, based on a semiconductor chip. The Proton I semiconductor chip is filled with millions of sensors. I don't have exact figures of how many sensors does an individual chip possess. But, I gather from reliable sources that the number is the Proton I semiconductor chip endorsed 165 million sensors, and the proton II 660 million sensors. The sensors are developed based on complementary metal-oxide semiconductor (CMOS) technology.
CMOS is actually a technology used in the manufacturing computer microchips, usually engineered from two semiconductor metals- Silicon and Germanium. The same technology is used in digital cameras, but here instead of sensing light, this sensor detects change in pH. What's a pH meter got to do here? The technology used here makes use of a simple principle in natural DNA replication. Each time a nucleotide is successfully incorporated into a growing DNA strand a hydrogen ion is released. For people, who are wondering where did this proton come from, Nucleic acids are acidic because of the phosphate groups. They can act as hydrogen ion donors, and proton is thrown out during nucleotide incorporation.
 |
Photo 2: Semi conductor chip
|
So, take a human genome, prepare a DNA library, add it on to the sequencing chip and each sensor acts as a well for reaction. When flooded with nucleotides the correct match is taken up and a change in pH is recorded which is converted to digital data. A powerful computer program integrates all the data and bingo, you have the sequence.
If I had to say anything more about this technology, I would say it works pretty much the same as the SMRT did. But the difference lies in the method of detection. Here the detection is based on pH change.
The battle to make it to the top is in between the Ion sequencer and its rival Illumina was so evident. Just a few days before the proton was announced HiSeq 2500 was released. There isn't enough conclusive evidence on which is actually better, the scientific community is more inclined to the ion proton version as it costs less ($740,000 vs $150,000). As a sequencer at a smaller scale, a modified version known as Ion Personal Genome Machine™ (PGM™) Sequencer competes with a illumina mini version (MiSeq). Again cost is an important factor ($50,000 vs $100,000 per machine). (Source)
"DNA sequencing is going to affect everything," says Rothberg, predicting it will become a $100 billion industry. "This is biology's century, just as physics was the foundation of the last century." (Source). And they also argue it as better than other technologies like Nanopore sequencing (Link).
Illumina sequencer
 |
Fig 5: dNTP used in SBS
|
Next let's talk something about the llumina/Solexa sequencer. The technology of sequencing is called as "Sequencing by synthesis technology". This technology is a brain child of 2 scientists from Cambridge- Shankar Balasubramanian and David Klenerman. They studied the movement of polymerase enzyme by using fluorescent dye labelled nucleotides. Based on their prime experience in sequencing project and their studies on polymerase, they theorized a massive parallel sequencing of short reads using a solid phase sequencing with reversible terminators as the basis of a new DNA sequencing approach. this technique came to be known as SBS or "Sequencing by synthesis" technology. Over the years, the technology was developed and a successful Solexa prototype was launched as a commercial sequencing instrument. A detailed history can be found here.
So how does the technology work? The technology is very much similar to Sanger sequencing method. The difference lies in use of modified dNTP's with terminators. 3′-O-fluorophore-labeled nucleotides were synthesized and used as reversible terminators of DNA polymerization. This reversible terminator ensures that in one step, only one nucleotide can be incorporated. After the template is flooded with nucleotides and binding step is accomplished, the unincorporated reagents are washed away. The terminator chemical is equipped with a fluorescent tag that allow it to be detected by using specific detection cameras. Since only one type of fluorescent color is used the detection of 4 nucleotides use 4 separate tubes. The 2nd step is to remove the terminators using a chemical reaction. This means, there is a removal of fluorescent tag also and the cycle is repeated.
The technology claims to be capable of a read length of nearly 50 bases for fragment libraries and 36 bases for mate-paired libraries, with a raw base-calling accuracy of 98.5% (Source is most probably outdated. I couldn't find the latest).
Of course there are more varieties coming up in the field of research and some new techniques are coming up in test mode. But I believe the post has give you a basic idea on some of the sequencer's that has now become the talk of DNA. A technology that is now gaining popularity is pore-fection or commonly known as nanopore sequencing. The technique is under development. Please refer to my previous post on pore-fection for more information on the technique (Link). The technique is emerging as potent, cost effective technology. Probably in a few years we will have machines that will sequence the human genome in less than an hour for less than 100$. And that will be a true genetic age of Lab science.
- Chun-Xiao Song etal. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nature Methods 9, 75–77. doi:10.1038/nmeth.1779. Link
- Paul Zhu and Harold G. Craighead. Zero-Mode Waveguides for Single-Molecule Analysis. Annual Review of Biophysics. June 2012; Vol . 41: 269-293. doi: 10.1146/annurev-biophys-050511-102338. Link
- Christian Castro etal.Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. PNAS March 13, 2007 vol. 104no. 11 4267-4272. Link
- Democratizing DNA sequencing by reducing time, cost and informatics bottleneck. Link
- aek-Soo Kim etal. Novel 3′-O-Fluorescently Modified Nucleotides for Reversible Termination of DNA Synthesis. ChemBioChem; January 4, 2010. Volume 11, Issue 1, pages 75–78. Link
- Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT (2012) Direct Comparisons of Illumina vs. Roche 454 Sequencing Technologies on the Same Microbial Community DNA Sample. PLoS ONE 7(2): e30087. doi:10.1371/journal.pone.0030087.



Comments
Post a Comment