Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from May, 2015

Posted by
Varun C N
Lab Series# 6: A short introduction to TALEN
- Get link
- X
- Other Apps

Posted by
Varun C N
Snippet: T cell exhaustion in HIV
- Get link
- X
- Other Apps

Posted by
Varun C N
What is not known about Rabies infection
- Get link
- X
- Other Apps

Posted by
Varun C N
Lab Series# 5: DNA amplification in Lab
- Get link
- X
- Other Apps
Posted by
Varun C N
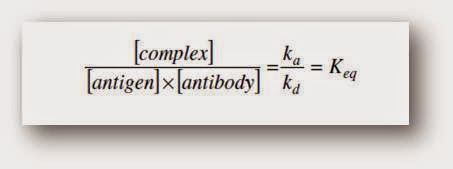
Lab Series# 4: Antigen Antibody reaction
- Get link
- X
- Other Apps