Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from September, 2015

Posted by
Varun C N
Lab Series# 7: O&P examination of Stool Samples
- Get link
- X
- Other Apps

Posted by
Varun C N
Wanted suspect: L pneumophilia
- Get link
- X
- Other Apps
Posted by
Varun C N
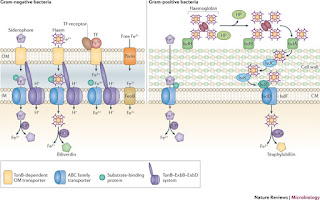
Will go to war for Iron: Bacterial strategy
- Get link
- X
- Other Apps

Posted by
Varun C N
BtB#4- Sterilization/Disinfection in brief
- Get link
- X
- Other Apps

Posted by
Varun C N
Ebola 2014: Not yet over in 2015!!
- Get link
- X
- Other Apps

Posted by
Varun C N
Blogger's Desk #8- Statistics in Layman's language
- Get link
- X
- Other Apps