Human Blood Virome
There is no denying that blood transfusion is an important part of modern medicine. In a good number of cases, the whole blood need not be given but rather a component of blood can be given. This allows the same blood to be given as components to different recipients as required. The ability to transfuse blood or its components is lifesaving in most cases. Bloodborne infections such as HIV, Hepatitis B and C etc. are routinely tested in the lab before the transfusion. As per the current guidelines, blood and blood-derived products (BBDP) are tested for a set of microbial agents which are thought to be pathogenic and transfusion- transfusible.
There is a debate on if we are testing enough. For example, testing strategies currently use an immunology-based assay for detection of signatures related to infections and that is not as sensitive as genetic tests. There are many other region specific infections such as Ebola and Zika virus (especially since certain carriers are asymptomatic) which can be transmitted by blood for which routine testing is not done. We are not even aware of every possible range of organisms that can be transmitted, let alone test. In an ideal case, the BBDP should be free of any microbial component. However, such a scenario is unlikely. Just as many other body fluids have been shown to be non-sterile, blood is also shown to have a good number of microbiome associated with it.
Most of the microbiome papers are actually bacteriome papers. Bacteriome papers are much more common in literature since it involves sequencing of 16s rRNA and it is much easier to do. In similar lines, mycobiome can be done, since it involves sequencing a particular target. In contrast, there is no common targetable gene or region of the genome for viruses and the only viable approach for studying virome is to do whole genome sequencing. Human virome has been published heavily for skin and gut and the major findings have been bacteriophages of various types. Blood virome is thus an interesting study that looks into what viruses are there in blood and are we testing enough.
In 2003, a proposal was made to develop a global system to catalog viruses and detect emerging diseases throughout the world. The idea was to routinely screen human blood to identify and monitor viruses from human samples. The core of the proposal was to collect blood from hospitals and labs weekly. This would be followed by extraction and sequencing of the viral genomes. Once a database of viruses is constructed, researchers could use it to screen for new viruses as they appear in the population. Such a database was supposed to help in quick identification of emerging infections and identify novel viruses.
A multi-group collaboration involving Dr. J Craig Venter (recipient of De Leeuwenhoek Medal 2015) who has been recognized for developing several generic findings by sequencing technologies has now published a paper on human blood virome from Human Longevity Inc. To put the study in a nutshell they sequenced genomes of 8,240 individuals who were all essentially healthy and not infected with anything. The majority of the reads were mapped to the human reference genome. Roughly 0.2% was attributed to bacteria and 0.01% to viruses. Fig 1 shows a summary of the percentage of individuals presenting with viral sequences. It was not surprising to see a variety of human herpes virus, but there were many other viruses some of which are related to humans and many bacteriophages. It must be noted that this study was designed to look for DNA viruses and there would be much more in the data if RNA-related virome reads can be obtained.
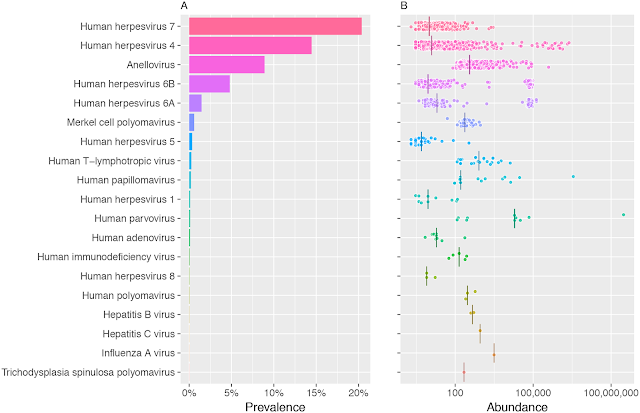 |
Fig 1: Prevalence and abundance of human DNA viruses and retroviruses in 8,240 individuals. A) Frequency B) The viral load. Source
|
Reference
Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y et al. The blood DNA virome in 8,000 humans. PLOS Pathogens. 2017;13(3):e1006292.





Comments
Post a Comment