MCR gene variants
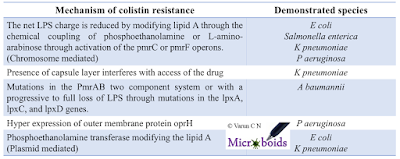 |
| Table 1: Known mechanisms of colistin resistance. |
MCR-1 a plasmid mediated resistance gene has been extensively discussed on this blog previously. Ever since its first report, a lot of research papers have come from the topic. There is hardly a country where genomic surveillance tools are available and MCR-1 is not reported. The location of the gene on a plasmid makes it more devastating. It is indeed very interesting to note that the MCR-1 is not just located in one plasmid. Many different plasmids have been demonstrated to be capable of harbouring MCR-1 and hence it should be inferred that it is widely distributed. It should be noted that the colistin resistance can be caused by several factors. Table 1 is a compiled list of colistin resistance determinants.
 |
| Fig 1: Structure of the catalytic domain of MCR-1 phosphoethanolamine transferase. Source |
The crystal structure of MCR-1 is already published and a good lot of information is available about the domain. The crystal structure study showed that MCR-1 possess a α-β-α-sandwiched structure and coordinate divalent zinc ions in the active site via phosphorylation of the conserved residue threonine. The nucleophile for catalysis is threonine 285 (Phosphorylated). They are most homologous to LptA and EptC transferases in terms of the sequence.
What is most interesting is that MCR variants are now being described. In a surveillance study at Belgium, 92 porcine and bovine colistin-resistant E coli isolates that were negative for MCR-1 was identified. Of these randomly selected 10 isolates, plasmid sequencing was done and 3 of the 10 showed sequence later designated as MCR-2 gene, which was 1,617 bp long, nine bases shorter than mcr-1(1,626 bp), and showed 76.75% identity. In 2016 august, a group from Italy described a novel MCR variant, named MCR-1.2, from an MDR KPC-producing K pneumoniae strain belonging to sequence type 512. This strain (KP-6884) was isolated in 2014 from a rectal surveillance swab obtained from an Italian child admitted to the paediatric onco hematology ward of Pisa University Hospital.
 |
| Fig 2: Sequence comparison of MCR and its variants. Source |
Most recently another MCR-1 gene variant, referred as MCR-1.3 carried is reported from a colistin-resistant S. Typhimurium strain YL14P053, which was isolated in 2014 from a rectal swab from a 46 years old healthy woman who received a medical examination in the Yulin Center for Disease Control and Prevention. It is a little confusing to me that the news and press release article mentions this variant as an MCR 1.6 whereas in the paper it says MCR 1.3. Fig 2 shows the alignment of MCR-1 gene with its variants are shown. MCR-1.2 has a variation at the nucleotide position 8, A was mutated to T, leading to a Gln to Leu change in amino acid position 3; MCR-1.3 has a variation in nucleotide position 1263, G was mutated to A, which is a synonymous mutation, while in nucleotide position 1607, G was mutated to A, leading to an Arg to His change in amino acid position 536.
Corresponding author Biao Kan comments, "This is the first time a mcr-1 gene has been found in Salmonella in a healthy carrier. Healthy carriers play an important role in the transmission of resistance genes to the community. Salmonella infections have been the leading cause of foodborne illness, and Salmonella-carrying mcr-1 will likely be a problem in food safety".
It occurs to me that the MCR gene and its variants have been found in several scenarios where there is no colistin pressure. That means evolutionarily there is no great fitness cost in keeping it, further suggesting that losing this plasmid would be a common feature once acquired.
Just like there are several NDM types and variants after the original discovery of NDM-1, am sure we are going to have several types of MCR-2. If we start whole genome sequencing of every colistin resistant isolate we are going to definitely get a lot of variants and maybe more types. Or who knows, something totally novel.
References
Olaitan A, Morand S, Rolain J. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Frontiers in Microbiology. 2014;5.
Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R et al. Diversifiedmcr-1-Harbouring Plasmid Reservoirs Confer Resistance to Colistin in Human Gut Microbiota. mBio. 2016;7(2):e00177-16.
Stojanoski V, Sankaran B, Prasad B, Poirel L, Nordmann P, Palzkill T. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biology. 2016;14(1).
Hu M, Guo J, Cheng Q, Yang Z, Chan E, Chen S et al. Crystal Structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin Resistance. Scientific Reports. 2016;6(1).
Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. Identification of a novel plasmid-mediated colistin-resistance gene, MCR- 2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27):pii=30280
Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S et al. mcr-1.2, a New mcr Variant Carried on a Transferable Plasmid from a Colistin-Resistant KPC Carbapenemase-Producing Klebsiella pneumoniae Strain of Sequence Type 512. Antimicrobial Agents and Chemotherapy. 2016;60(9):5612-5615.
Lu X, Hu Y, Luo M, Zhou H, Wang X, Du Y et al. MCR-1.3: a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolated from a healthy individual. Antimicrobial Agents and Chemotherapy. 2017;:AAC.02632-16.





Comments
Post a Comment