Chip makes a quick decision about antibiotics
Greetings
Let me revisit the topic of antibiotic resistance. Antibiotic misuse is a leading cause of resistance. The misuse arises due to two reasons- unwarranted broad spectrum antibiotic prescription and unregulated use There is a lot of literature reporting the inappropriate use. This stems from having to guess the organism and start therapy before the lab tests are back. Guessing the organism is probably the easier part but predicting the antibiotic is more difficult. This has been eased to an extent by surveillance studies that identify the local antibiogram pattern. However, given the evolving nature of bacteria many a times the best guess is not even near good. There are several important questions and concepts when it comes to use of antibiotics. I recommend you read a blog post by Sridhar Rao
This is a standard clinical problem in most parts of the world. The problem arises from the need to grow the organism for testing. An alternative is genomic or proteomic based testing. These methods are expensive. Though whole genomic sequencing and MALDI TOF instruments are pushing the boundaries its application is still not in wide use. Also use of such methods require that resistance signatures are already known in the database. This has pushed the need to bring in systems that are rapid, reliable and low cost. A new paper published in LabChip by Justin Besant and team looks into just that. As Besant comments, "Guessing can lead to resistance to these broad-spectrum antibiotics, and in the case of serious infections, to much worse outcomes for the patient, we wanted to determine whether bacteria are susceptible to a particular antibiotic, on a timescale of hours, not days."
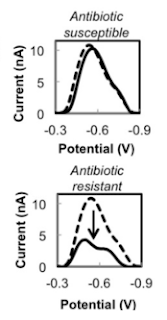 |
| Fig 2: Representative differential pulse voltammograms. Source |
What exactly is antibiotic resistance? You can get so many definitions based on which guidelines are talking about it (Seriously!!). But perhaps the definition that nails it reads so, "If organism is active metabolically in presence of the inhibiting agent, that's resistance". That means if you can demonstrate metabolism in organism, given sufficient time of exposure we can detect resistance. As mentioned in the paper there are several methods of achieving this under research such as detection of bacterial motion using AFM cantilevers, electrochemical measurements of bacterial growth, optical detection of bacterial growth, and optical detection of redox reporters of bacterial metabolism.
 |
| Fig 1: Schematic of in‐well bacterial capture. Source |
In the study the researcher's used a microfluidic chamber, containing microbeads trapping the bacteria from sample. This is trapped in the nano-well, containing antibiotic and resazurin. Resazurin is a proven redox indicator used in detecting bacterial metabolism. By comparing the electrochemical signature of resazurin, the test indirectly measures the metabolism and thus resistance. The test is extremely rapid owing to micro volumes and results are quite fast. Indeed the turn-around time was as short as 30 min (Not even genetic test is that fast
Of course this technique would require some additional tuning and standardization for routine microbiology tests. But I think this technique is going to make it to the market, as a benchtop automated analyzer. As the author puts it, "The electronics for our electro-chemical readout can easily fit in a very small benchtop instrument, and this is something you could see in a doctor's office, for example. The next step would be to create a device that would allow you to test many different antibiotics at many different concentrations, but we're not there yet."
Besant JD, Sargent EH, & Kelley SO (2015). Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab on a chip PMID: 26008802



Comments
Post a Comment