Teixobactin: Resistant to Resistance
Greetings,

Antibiotics have been such an important part of the current clinical practice, that it is unimaginable to see how we could do without it. With warnings of post antibiotic era fast approaching (Link), there is an attempt made to bring more chemicals in the list. The faster rate of antibiotic approval remains the same. Several new compounds have been approved for treatment. But the problem remains the same- Antibiotic resistance. I have previously blogged about why halting resistance to antibiotic doesn't look like a possibility (Link). However, the current talk on web pages is that a new chemical Teixobactin has pulled it off. Somebody called it resistance to antibiotic resistance.
There are 2 possible ways where you make antibiotic discovery. First, you synthesize chemicals unheard of in nature or a modified chemical from what already looks like a good antibiotic. Second is to tap into the microbial communities that have been producing antibiotics for self defense. But here's an important catch. More than a 99% of bacteria is unknown to science. All we know is they exist, through DNA sequencing approaches. These form a chunk of untapped resources. But to identify biology and chemicals of interest, from these we must first grow them. And they don't grow in laboratory conditions.
Apparently there is a dual problem of interest. The first part is how to culture the so called non cultivable bacteria, and the second using them to find chemicals of interest.
Unable to cultivate a huge set of microbes is a long standing problem. I had written in a post long ago (Link), that many of the organisms could grow in normal medium, provided they are given a chance to. The idea is that rapid grower's use up the nutrients in medium thereby hindering the slow growers. By eliminating the rapid growers, it was possible for slow growers to grow. Sometimes a more complex nutritional supply allows growth. I recently learnt from a post in ASM blog, the standard practices in microbial culture, for example use of agar in itself can be inhibitory to at least a select group of organisms. (Link)
But despite heroic efforts there seems to be a large percentage of bacteria that simply refuse to grow in a pure culture. The best example are soil microbes. It is believed that chemicals from one bacteria might somehow support the other and so probably they cannot grow in isolation, no matter how good the condition is. Thus study of these organisms have been hindered and the information they contain, is called as "Dark microbial matter".
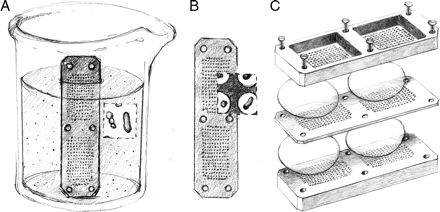 |
Fig 1: Isolation chip for microbial cultivation.
|
The technology to cultivate at least 50% of such non cultivable bacteria comes in the form a method called as Ichip (I chip stands for isolation chip, and its not an apple product). The Ichip is an insitu diffusion chamber. The basic design is composed of several hundred miniature diffusion chambers. By diluting the sample to a degree that will allow 1 microbial cell per chamber, it is possible to achieve isolation. On other hand since it is made up of a semi permeable membrane, it tricks the microbe to its natural environment, thereby inducing growth. There have been modifications of this original method, which are reported. For example, MicroDish Culture chip (MDCC) is a disposable microchip which can serve as a micro petri dish, The MDCC20 accommodates 1000 compartments . One more example that I can think of is SlipChip. Once a micro-colony can be obtained it is possible to grow them in larger numbers. That looks like a pretty good solution to a complicated problem.
The researchers using this knowledge, worked on trying to isolate soil microbiome. Extracts from 10,000 isolates obtained by growth through Ichip were further screened for antimicrobial activity against S aureus. During the screen they hit upon 25 potential chemicals. One of them was Teixobactin. The source organism was a β-proteobacteria provisionally named Eleftheria terrae. The researchers have done a lot of additional work to characterize the organisms biochemical pathways, entire genome and the chemical. However, it is not relevant for this post.
 |
| Fig 2: Teixobactin Structure. Source |
Teixobactin strongly inhibited synthesis of peptidoglycan hinting that it acts by attacking the cell wall. Experiments show unlike Penicillin class, these specifically interacts with the peptidoglycan precursors (decaprenyl-coupled lipid intermediates of peptidoglycan). The compound was exceptionally active against C difficile and Bacillus anthracis (MIC in ng/ml), and high activity against Streptococci, Enterococci and M tuberculosis (MIC <1 mcg/ml). The activity against TB is probably explained by the drug binding capabilities to precursors which are common for peptidogycan and arabinogalactan. However, they seem to be of no use against Gram negatives where the concern is higher.
 |
| Fig 3: Resistance acquisition. Source |
Drug resistance experiments, using serial sub MIC method, for 28 days in comparison with ofloxacin is strikingly distinct. There is almost no signal of resistance. This is the experimental for claims "Resistance to resistance". Its my opinion that Ofloxacin against which comparison is made is not a good way to do this experiment. The Resistance acquisition experiment should have been shown with vancomycin and daptomycin which would have been a better and crisper comparison. Moreover ofloxacin is a fluoroquinolone which acts through a totally different mechanism. Experiments have also been done to show that it is not toxic.
Teixobactin attacks lipids in cell wall, a highly fundamental part of the cell, because of which development of resistance can be slow. However, to call resistance will never emerge, is very difficult point to make. As Wright states in nature news "“I don’t believe there’s such a thing as an irresistible antibiotic, but I do believe that certain antibiotics have a low frequency of resistance". Moreover, this is a isolate from soil microbiome. A resistome is definitely out there.
Outside the purview of this post, I want to digress syaing there is a way of predicting resistance in-silico. This is a software - called OSPREY which predicts the most likely mutations that a bug develops in response to a new drug before the drug is even given to patients. It would be interesting to see if someone did the check to see what the software predicts for Teixobactin.
This paper will inspire more such studies alike and maybe will uncover a potential drug against the gram negatives. It is also intriguing to me what would have they found if they had screened the chemicals against say a MDR Klebsiella species. Someone needs to do that experiment to see if there are anti GNB's of interest. I'm sure there will be at least a handful of interesting candidates.
This paper will inspire more such studies alike and maybe will uncover a potential drug against the gram negatives. It is also intriguing to me what would have they found if they had screened the chemicals against say a MDR Klebsiella species. Someone needs to do that experiment to see if there are anti GNB's of interest. I'm sure there will be at least a handful of interesting candidates.
Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, & Epstein SS (2010). Use of ichip for high-throughput in situ cultivation of "uncultivable" microbial species. Applied and environmental microbiology, 76 (8), 445-50 PMID:20173072
Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, & Lewis K (2015). A new antibiotic kills pathogens without detectable resistance. Nature PMID:25561178




Comments
Post a Comment