Lab Series# 10: MALDI TOF in Microbial diagnostics
Greetings
I have many a times talked about the current issues with Clinical microbiology- rising antibiotic resistance and too much of time taken for pathogen identification. The new 4th generation technologies are aiming at rapid workup with very high sensitivity. There is a lot of research on reducing this large turn around time. One of the systems that has more recently come into the market is MALDI-TOF based microbiology identification. I have been personally running some tests that for more than a year now. For the 200th post in this blog site, I think its worth a good share.
Let me digress a little bit and talk about Mass Spectrometry (MS). The credits of inventing MS is partly owed to findings by Joseph John Thompson and Francis William Aston. The modern MS was developed by Arthur Jeffrey Dumpster.
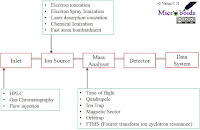 |
| Fig 1: Components of Mass Spectrometer. |
MS is an instrumental technique used to calculate mass to charge ratios (m/z) and relative abundances for any given molecule. In this case m stands for mass and z stands for charge number of ions. Consider you have two types of balls one heavier and the other lighter. Assume that the balls are travelling at a speed and you have a strong air flowing in a perpendicular path. The lighter is the ball, easier is it to deflect its path. In essence, if you know the original path, speed and the force used for deflection, you can calculate the mass of the ball. That's pretty much how the classic mass spec works.
An illustration of general outline of MS is shown in Fig 1. For different steps many different variants are available, of which most commonly used versions are shown. MALDI TOF is a build where the ionisation process is through matrix Laser desorption and detection is through time of flight. Hence the name- Matrix assisted laser desorption/ionisation- time of flight, or in short MALDI TOF. In the original studies, it was found that alanine could be ionised when mixed with tryptophan and irradiated with a pulsed 266 nm laser. It was identified with subsequent studies that there are better matrix which can confer better ionisation properties. Examples include- Cinnamic acid derivatives, Picolinic acid etc. In this case, the time taken for the molecule to reach the detector is calculated as TOF, which functions as the mass analyser. The m/z values generated is plotted as a graph.
 |
| Fig 2: MALDI TOF Spectra analysis |
The trick in identifying the organism is to release the abundant protein such as Ribosomal proteins which have unique signatures for the species. With large scale studies a database has been generated containing m/z values for multiple species. The obtained sample plot is compared with the database and an algorithm calculates the closest possible match. An example of Mass spec comparison is shown in Fig 2. The upper part of the graph shows the spectra generated from my sample and the lower part from the spectra from database. For a closer look I have shown an example of detailed spectra in the higher molecular range. The red lines indicate the unmatched m/z values and green indicates good match. Depending on number of m/z ratio's matched the confidence in the identification varies. If database exists for strain subtype and similar peaks can be found in the generated spectra, strain typing can be directly done. That means there is an inherent limitation- "The database".
 |
| Fig 3: MALDI TOF Process |
For most of the bacterial isolates, obtained in fresh culture the proteins are easily release and ionised. The cultured bacteria is spotted on a MALDI plate, to which formic acid and matrix is added (I used α-Cyano-4-hydroxycinnamic acid reagent). The spots are bombarded with high energy laser. A representation of the process is shown in Fig 3. For more difficult organisms such as Mycobacterium species, a extraction step is recommended (I have tried with Acetonitrile extraction method).
 |
Photo: I and Dr. Shivaram working
on MALDI TOF MS.
|
The MALDI TOF systems are used in multiple laboratories worldwide. The system allows identification of organism in less than 10 min. More recently techniques have been developed where the organism can be analysed directly from sample. For example in case of bacteraemia, the blood sample is treated with a reagent that reduces host proteins to a very large extent. Subsequently, MALDI TOF can be run with extract. Within an hour the results can be obtained. That is a huge leap from conventional method.
With help of MALDI TOF, bacterial and fungal strains are well identified. As I already specified, the method is as good as the database is. Identification of virus would be an obvious target. However, most viruses have very few proteins. Additionally, they can't be independently cultivated. Hence, always there are a lot of other proteins in background which will mask the very few viral proteins. Despite the problems, attempts have been made to identify viral strain, with a small success percentage.
The next challenge is antibiotic sensitivity. MALDI TOF MS inherently detects and creates a spectra ID for anything that you feed it. Antibiotic resistance can be due to a huge combination of protein changes and a database for every variant is seriously beyond the scope at least for now. But there is a possible alternative method for detecting some types of resistance. For example, if the system can show that a drug is being cleaved by the isolate then it must be resistant, irrespective of its changes in its proteins. An example of this is Meropenem hydrolysis assay. Strain is incubated with meropenem and MALDI TOF is run for the mixture. By looking for peaks which correlate with cleavage products of Meropenem, carbapenemase resistance can be indirectly inferred. The time of work can be reduced to about 3-4 hours. Now studies are also showing this could be done from sample directly.
Overall, MALDI TOF is a great method especially when Turn around time needs to be as fast as it can be.
Murray PR (2012). What is new in clinical microbiology-microbial identification by MALDI-TOF mass spectrometry: a paper from the 2011 William Beaumont Hospital Symposium on molecular pathology. The Journal of molecular diagnostics : JMD, 14 (5), 419-23 PMID: 22795961
Biswas S, & Rolain JM (2013). Use of MALDI-TOF mass spectrometry for identification of bacteria that are difficult to culture. Journal of microbiological methods, 92 (1), 14-24 PMID: 23154044
Hrabák J (2015). Detection of carbapenemases using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) meropenem hydrolysis assay. Methods in molecular biology, 1237, 91-6 PMID: 25319782





Comments
Post a Comment