Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from October, 2016

Posted by
Varun C N
ETX2514: A new Beta Lactamase Inhibitor
- Get link
- Other Apps
Posted by
Varun C N
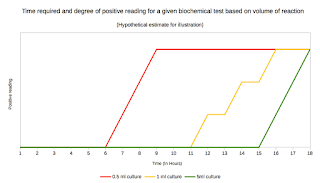
Lab series# 15: Biochemical tests for identification of bacterial isolates
- Get link
- Other Apps

Posted by
Varun C N
Lyme disease: A possible link to Swiss agent?
- Get link
- Other Apps
Posted by
Varun C N
Understanding Autophagy: Nobel winning concept
- Get link
- Other Apps

Posted by
Varun C N
Rhinovirus C
- Get link
- Other Apps

Posted by
Varun C N
Nobel Awards- 2016
- Get link
- Other Apps