Microbiome- Dwelling a little deep
Greetings
For every bacterial species known, there are bacteriophages. Gut microbiome is perhaps the most heavily colonized microbiome. with more than 35000 varieties of bacteria in the site, I expect at least a million (if not billion) phages there. The best part is they form a defense layer. Innate immunity acquired via such phages are referred as, phage-mediated immunity (PMI). Study by Barr etal, had shown that our mucus membrane is heavily colonized by lytic phages, which tend to attack incoming invaders and serve as first line of defense. Due to their extreme importance in defense, there is a expected evolutionary pressure to retain these phages. It is not known as to how these phages are maintained, or if at all it is maintained by the host or the gut microbiota. As the next story will show bacteria can equip themselves with more than toxins to counter other invading pathogen. They can arm themselves with phages against others. So maybe, the microbiome maintains the phages. After all its the requirement of normal flora to stay in competition for their own benefit.
Let me digress a little bit, to cite maintenance of virus as an armory. This is based on an article published by Lora Hooper and team. The study showed that Enterococcus faecalis has in its genome integrated genomes of two different phage elements (one gives structure to the composite, while the other helps it infect victims), which can be unleashed on demand. These phages attacks other bacteria providing a competing advantage. This strain is called V583. That's a proof of concept, and maybe PMI is replenished through similar mechanism. I however, have no data to support my idea.
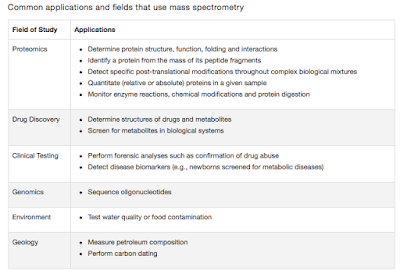
Everyone is familiar with the nomenclature "Normal flora". They represent a set of microbes that inhabit us. Estimates have claimed that there are nearly 10 times more microbes in human body than the cells. Debates exist as to if this is true or an exaggeration. But it is irrelevant at this point. It is for sure however that they interact and influence our cellular machinery. Their influence is studied in terms of Immunological signatures, neurobiology, oncogenesis etc. Most of the textbooks don't acknowledge this yet, since the details are just beginning to emerge. I have blogged about these aspects a variety of time in this blog which you could check out.
 |
| Fig 1: A representation of common members of bacteriome. Source |
I have been thinking for sometime, "Is it worth to call our microbiome a yet another organ"?. I haven't posed this question for the first time. This question has been discussed in great depth in a variety of conferences, academic papers etc. What is not realized is the human microbiome is much more than that, and its dynamic is too difficult to study. The term microbiome in itself is considered as a sloppy definition (Read ASM blog post here). For the purpose of clarity, let us stick to the definition that microbiome denote all microbes at a site. The concept of microbiome is derived from the development of deep sequencing methods with billions of reads. Even the best of microbial culture method cannot retrieve and analyse every possible species in a site. Clearly we had to await the emergence of modern technology. A massive Human microbiome project, is currently pursuing studies to understand the human microbiome. It is clear that microbiome is a signature of individual and there is a huge variation. Its uniqueness is so striking that it has been proposed as a forensic tool. Body sites presumed to be sterile in healthy humans, such as the vascular endothelium, Brain etc have been shown to be colonized without apparent signs of disease. Such publications have shaken the ground beliefs of sterile sites in body.
Till extremely recently the word microbiome was almost exclusively studies on bacteriome. This is because, almost all bacteria have a signature identifiable sequence in one single gene- 16S ribosomal sequence. Amplify and identify all the 16S deep sequences (not as simple as said), a metagenomic approach and boom you have everything. Fungal microbiome is a little more harder and the virome is the hardest. As represented in Fig 2, expected virome content is much more than bacteriome or mycobiome. For virome, you just deep sequence everything you find and remove the known bacteriala nd other DNA matches. The rest is matched directly with known database and similar looking viral genomes from database. Often there will be "grey sequences" (casually referred as dark matter) which doesn't match with anything.
 |
| Fig 2: A rough representation of microbiome content. |
So much of information to bring you into a question, one of the readers had asked me after reading my previous post on "Algal virus and cognition". Is there examples of good or bad effects of human virome? And suddenly I realized that I have never talked much about the Human Virome.
As can be understood from above, "microbiome" is a massive subject of interest with too much grey area to even speculate on its functionality. As I have said, bacteria has been studied more in connection with a variety of conditions. But it simply seems logical to me, to extend that where there is a bacteria, viruses will tag along. But the current virome literature is in its infancy and so I chose to put up some examples taken from studies. This should help in getting the answer across allowing room for exploring.
 |
| Box 1: Projected contents of human virome. |
Let me digress a little bit, to cite maintenance of virus as an armory. This is based on an article published by Lora Hooper and team. The study showed that Enterococcus faecalis has in its genome integrated genomes of two different phage elements (one gives structure to the composite, while the other helps it infect victims), which can be unleashed on demand. These phages attacks other bacteria providing a competing advantage. This strain is called V583. That's a proof of concept, and maybe PMI is replenished through similar mechanism. I however, have no data to support my idea.
I have talked about HERV in a post almost about 2 years ago emphasizing the fact that our very existence as to humans owes a lot to these viral pieces. Just to remind, the placental formation is based on a viral protein (Syncytium beta)- HERV-W envelope glycoprotein coding gene. Read my previous post here. Many more such marvels are slowly being discovered. As explained in a previous post (Link), Inactively residing herpes in neurons may have beneficial effects.
Just as the bacteriome influences outcome in many different conditions so does viruses. Studies are currently studying viral composition in a variety of disease conditions. One of the most cited study is by Moya etal; showed there is less viral diversity in the Crohn’s disease patients compared to healthy. Additionally they found these where mostly Retroviridae members in effected patients. My take on this there is a lack of phage varieties that effect the bacteriome. It is known that there is a significant importance of bacteriome quality and type in crohn's disease.
The said field offers new research avenues, but comes with its own set of challenges. Microbiomes are not stagnant types in a person. At least some percentage of it vary continuously. By using a CRISPR based tagging approach, in a 11 months study it was shown that the virome can vary by anywhere between 25 and 75% between every sample. This means that it is computationally and logically difficult to characterize a virome and associate it with a condition.
Just to add to flavor there is a realm of fungus in the microbiome. Recently published data suggests that fungus too have dynamic interaction. A study has gone as far as to characterize how fungal microbiome can dictate T cell balances (which can be done by bacteria and some helminths as well), thereby creating a risk for allergies. And then there are phages of fungus called as mycophages which am sure will be present as a part of our virome. The entire field of mycophages is currently a dark region, so forget about commenting on its effects.
To conclude, I will say microbiome is currently a hot topic. Connection has been made with a variety of conditions and microbiome. But proving and characterizing every connection is very hard at least with current technology. Virome is a step next to it, though science will make its advance slowly.
The said field offers new research avenues, but comes with its own set of challenges. Microbiomes are not stagnant types in a person. At least some percentage of it vary continuously. By using a CRISPR based tagging approach, in a 11 months study it was shown that the virome can vary by anywhere between 25 and 75% between every sample. This means that it is computationally and logically difficult to characterize a virome and associate it with a condition.
Just to add to flavor there is a realm of fungus in the microbiome. Recently published data suggests that fungus too have dynamic interaction. A study has gone as far as to characterize how fungal microbiome can dictate T cell balances (which can be done by bacteria and some helminths as well), thereby creating a risk for allergies. And then there are phages of fungus called as mycophages which am sure will be present as a part of our virome. The entire field of mycophages is currently a dark region, so forget about commenting on its effects.
To conclude, I will say microbiome is currently a hot topic. Connection has been made with a variety of conditions and microbiome. But proving and characterizing every connection is very hard at least with current technology. Virome is a step next to it, though science will make its advance slowly.
Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vázquez-Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, & Gilbert JA (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science, 345 (6200), 1048-52 PMID:25170151
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, & Knight R (2009). Bacterial community variation in human body habitats across space and time. Science, 326 (5960), 1694-7 PMID:19892944
Wylie KM, Weinstock GM, & Storch GA (2012). Emerging view of the human virome. Translational research : The journal of laboratory and clinical medicine, 160 (4), 283-90 PMID:22683423
Barr JJ, Youle M, & Rohwer F (2013). Innate and acquired bacteriophage-mediated immunity. Bacteriophage, 3 (3) PMID: 24228227
Pérez-Brocal V, García-López R, Vázquez-Castellanos JF, Nos P, Beltrán B, Latorre A, & Moya A (2013). Study of the viral and microbial communities associated with Crohn's disease: a metagenomic approach. Clinical and translational gastroenterology, 4 PMID: 23760301
Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Núñez G, & Shibuya A (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE₂. Cell host & microbe, 15 (1), 95-102 PMID: 24439901



Comments
Post a Comment