Quasi Species- Discussion
Greetings
I have been posting regularly on "Infection Outbreaks". In my previous post, I talked about Indian Influenza outbreak that has been concerning and received some attention. With a publication claiming there is a mutation of interest which may make the H1N1 more pathogenic but Indian studies showing that there isn't such a scenario has created a duality (Link). But what causes emergence of Rapid mutation in viruses like that of Influenza? That's an interesting question to ask. May be the question deserves a blog space here. I have previously talked about mutation (Link here and here).
Let us take the example of Q-beta phage which first demonstrated the depth of genetic variance in virus. Let me quote from a post in virology blog (Link). "A Q-beta phage population is in a dynamic equilibrium with viral mutants arising at a high rate on the one hand, and being strongly selected against on the other. The genome of Q-beta cannot be described as a defined unique structure, but rather as a weighted average of a large number of different individual sequences". This dynamism in the virus population within the species. There is misinterpretation that virus have a excessive mutation rate. It is true that there is about one magnitude difference in mutation rate for viral genomes, thanks to the lower fidelity of viral replication enzyme. But its the massive replication that leads the way.
Let me cite Influenza as an example, since it is of great interest. In experimental conditions, Influenza A virus begins producing a progeny aprox 6 h after infection of cell, and continue to do so for another 5 h. A single infected cell produces 22 new productive infections on average. Mathematically, even if you had consider that there is a mutation once in a 1000 replication, at the end of a week, you would have enough genetically diverse types. You also have to consider fitness cost. Not all mutations are good for the virus to have. So many of them would simply vanish of. So, if you had take a sample of viral population in circulation, it is almost imperative that you had find some mutations. To have a clear idea of how much the mutation has penetrated, you need to have lot of sequences.
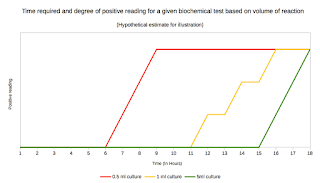
 This concept presents with an often overlooked point. For many viral infections seroconversions and rapid accumulation of mutation is a common phenomenon. This doesn't mean things suddenly happened that way overnight. For example, Ebola is a case of recent interest. In ever case there would be a large set of viruses that have slightly different mutation. However, only the one's which resist the immune attack and are successful would be predominantly present in the sample (Fitness cost), and studies will yield the same. That's the whole point of constructing dendrograms. It lets you elute the dynamism. I recently had a discussion about this, and I thought its worth posting the same here. See Fig 1
This concept presents with an often overlooked point. For many viral infections seroconversions and rapid accumulation of mutation is a common phenomenon. This doesn't mean things suddenly happened that way overnight. For example, Ebola is a case of recent interest. In ever case there would be a large set of viruses that have slightly different mutation. However, only the one's which resist the immune attack and are successful would be predominantly present in the sample (Fitness cost), and studies will yield the same. That's the whole point of constructing dendrograms. It lets you elute the dynamism. I recently had a discussion about this, and I thought its worth posting the same here. See Fig 1
 This concept presents with an often overlooked point. For many viral infections seroconversions and rapid accumulation of mutation is a common phenomenon. This doesn't mean things suddenly happened that way overnight. For example, Ebola is a case of recent interest. In ever case there would be a large set of viruses that have slightly different mutation. However, only the one's which resist the immune attack and are successful would be predominantly present in the sample (Fitness cost), and studies will yield the same. That's the whole point of constructing dendrograms. It lets you elute the dynamism. I recently had a discussion about this, and I thought its worth posting the same here. See Fig 1
This concept presents with an often overlooked point. For many viral infections seroconversions and rapid accumulation of mutation is a common phenomenon. This doesn't mean things suddenly happened that way overnight. For example, Ebola is a case of recent interest. In ever case there would be a large set of viruses that have slightly different mutation. However, only the one's which resist the immune attack and are successful would be predominantly present in the sample (Fitness cost), and studies will yield the same. That's the whole point of constructing dendrograms. It lets you elute the dynamism. I recently had a discussion about this, and I thought its worth posting the same here. See Fig 1
Baccam, P., Beauchemin, C., Macken, C., Hayden, F., & Perelson, A. (2006). Kinetics of Influenza A Virus Infection in Humans Journal of Virology, 80 (15), 7590-7599 DOI: 10.1128/JVI.01623-05
Nobusawa, E., & Sato, K. (2006). Comparison of the Mutation Rates of Human Influenza A and B Viruses Journal of Virology, 80 (7), 3675-3678 DOI: 10.1128/JVI.80.7.3675-3678.2006
Sanjuan, R., Nebot, M., Chirico, N., Mansky, L., & Belshaw, R. (2010). Viral Mutation Rates Journal of Virology, 84 (19), 9733-9748 DOI: 10.1128/JVI.00694-10



Comments
Post a Comment